"These breakthroughs introduce us to a golden era of science and completely rewrite our future."
Demis Hassabis, CEO of DeepMind, junior chess prodigy, former game developer and, of course, UCL alumnus, made headlines last week by joining the ranks of Nobel Laureates in Chemistry. He and his colleague, John M. Jumper, senior science researcher at DeepMind, achieved a game-changing breakthrough by pioneering new methods for predicting protein structures with Artificial Intelligence, and were awarded the half the Nobel Prize for their efforts.
Along with that, David Baker, professor of biochemistry and Director of the Institute for Protein Design at the University of Washington has been awarded the other half of the 2024 Nobel Prize in Chemistry for computational protein design.
These breakthroughs introduce us to a golden era of science and completely rewrite our future. And here's why we must talk about it.
David Baker
“In nature, proteins are the miniature machines that carry out all the important jobs: we can think, we can move, we can digest food, plants can capture energy from the sunlight and everything that happens in the living organism is due to proteins. We can use proteins to solve problems that evolution did not manage to solve.” - David Baker.
Each protein chain folds into its own characteristic shape and the folding process is very precise. The shape of a folded protein chain is what defines its biological functions. However, there are so many different shapes a protein can adopt, which made the protein problem so difficult and rendered it unsolved for over 50 years. Up until now.
The gigantic increase in computing power since the problem’s discovery now enables us to design tens of thousands of new proteins with new shapes and new functions. There are now over 10130 different designs we can explore using computation, enormously larger than the total number of proteins that have existed since life on earth began. After creation, we can extract the proteins, and then determine their functions and whether they are safe.
Today we face challenges such as serious ecological threats as well as new diseases evolving, and we do not have millions of years to wait for the discovery of the right proteins. But using computational design tools, we can now build these completely new multi-purpose proteins.
John M. Jumper and Demis Hassabis
The AI system AlphaFold2 by DeepMind is the first non-experimental method that can predict the complex structure of any known protein in nature, also solving the "50-year grand challenge", in the words of Hassabis himself. This system can predict the way a protein folds based on the amino acid sequence it consists of, which enables us to create proteins that can perform very specific functions and help us drive humanity's development.
Until very recently it could take research biologists a year to get a single answer about a 3D shape of a protein fold; now we have a machine learning algorithm that can do the same in 5-10 minutes.
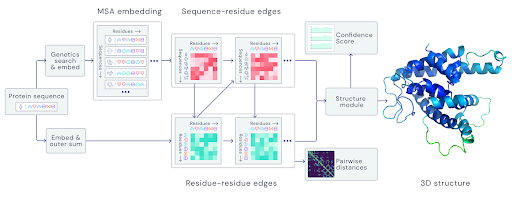
The program consists of three stages: database search and preprocessing, EvoFormer, and structure model.
Database search and preprocessing. A sequence of amino acids is entered, and AlphaFold compares it to records from several databases to extract similar sequences from other organisms. It also creates a pair representation of this input sequence, showing pairs of amino acid residues that are close together in 3D space within the target protein. Residue is the part of an amino acid that remains after it forms a peptide chain with other amino acids, and water is removed.
EvoFormer is a unique AlphaFold neural network that looks for relationships in residue pairs of the input sequence. It also evaluates the relationship within any two residues, which can be thought of as nodes in NNs. These calculations are carried out 48 times before forming a refined model of residue pair representations.
Finally, the Structure Model is another neural network that takes the previously refined model and performs rotation and translation on it to create a prediction of what its 3D protein structure looks like.
These newly developed proteins can fight cancer, break down plastic waste, and develop vaccines for respiratory diseases among many other uses. This intersection of biology and AI in 2024 can help us uncover the secrets of life, fight diseases, and even address the overwhelming climate challenges of today and the future.
